Simple MLP - Forward and Backward Pass (With Einsum) Derivation
Neural Network Forward and Backward Pass
Notation
h = Sigmoid(wx + b)w = Nh × NiV = No × Nhy' = Softmax(Vh + c)b = Nhc = No-
x = Ni × 1(1 training sample)
Forward Pass
\[\begin{aligned} h_a &= W @ x + b \quad && (Nh \times Ni) @ (Ni \times 1) + (Nh \times 1) \\ h &= \text{Sigmoid}(h_a) \quad && (Nh \times 1) \\ y_a &= V @ h + c \quad && (No \times Nh) \times (Nh \times 1) + (No \times 1) \\ y &= \text{Softmax}(y_a) \quad && (No \times 1) \end{aligned}\]Cross-entropy loss: \(L = -\sum_{i=0}^{n} t \log y_i \quad \text{(scalar)} \rightarrow -t \log y \text{(vector)} \quad (No \times 1)\)
Backward Pass
Gradients w.r.t. loss:
\[\frac{\partial L}{\partial L} = 1 ; \quad \frac{\partial L}{\partial y} = -\frac{t}{y}\] \[\frac{\partial L}{\partial y_a} = \frac{\partial L}{\partial y} \cdot \frac{\partial y}{\partial y_a} = y - t \quad (No \times 1)\]Note: Output shape must match the variable shape w.r.t. which derivative is taken.
Derivatives:
\[\frac{\partial L}{\partial V} = \frac{\partial L}{\partial y_a} \cdot \frac{\partial y_a}{\partial V} = (y - t) \cdot h^\top \quad \Rightarrow \quad (No \times 1) @ (1 \times Nh) = (No \times Nh)\] \[\frac{\partial L}{\partial h} = \frac{\partial L}{\partial y_a} \cdot \frac{\partial y_a}{\partial h} = V^\top \cdot (y - t) \quad \Rightarrow \quad (Nh \times No) @ (No \times 1) = (Nh \times 1)\] \[\frac{\partial L}{\partial c} = \frac{\partial L}{\partial y_a} \cdot \frac{\partial y_a}{\partial c} = (y - t) \cdot 1 \quad \Rightarrow \quad (No \times 1)\] \[\frac{\partial L}{\partial h_a} = \frac{\partial L}{\partial h} \cdot \frac{\partial h}{\partial h_a} = \frac{\partial L}{\partial h} \left[ \sigma(h_a) \cdot (1 - \sigma(h_a)) \right] \quad \Rightarrow \quad (Nh \times 1) \cdot (Nh \times 1) = (Nh \times 1)\] \[\frac{\partial L}{\partial w} = \frac{\partial L}{\partial h_a} \cdot \frac{\partial h_a}{\partial w} = \frac{\partial L}{\partial h_a} \cdot x^\top = (Nh \times 1) @ (1 \times Ni) = (Nh \times Ni)\] \[\frac{\partial L}{\partial b} = \frac{\partial L}{\partial h_a} \cdot \frac{\partial h_a}{\partial b} = \frac{\partial L}{\partial h_a} \cdot 1 = (Nh \times 1)\]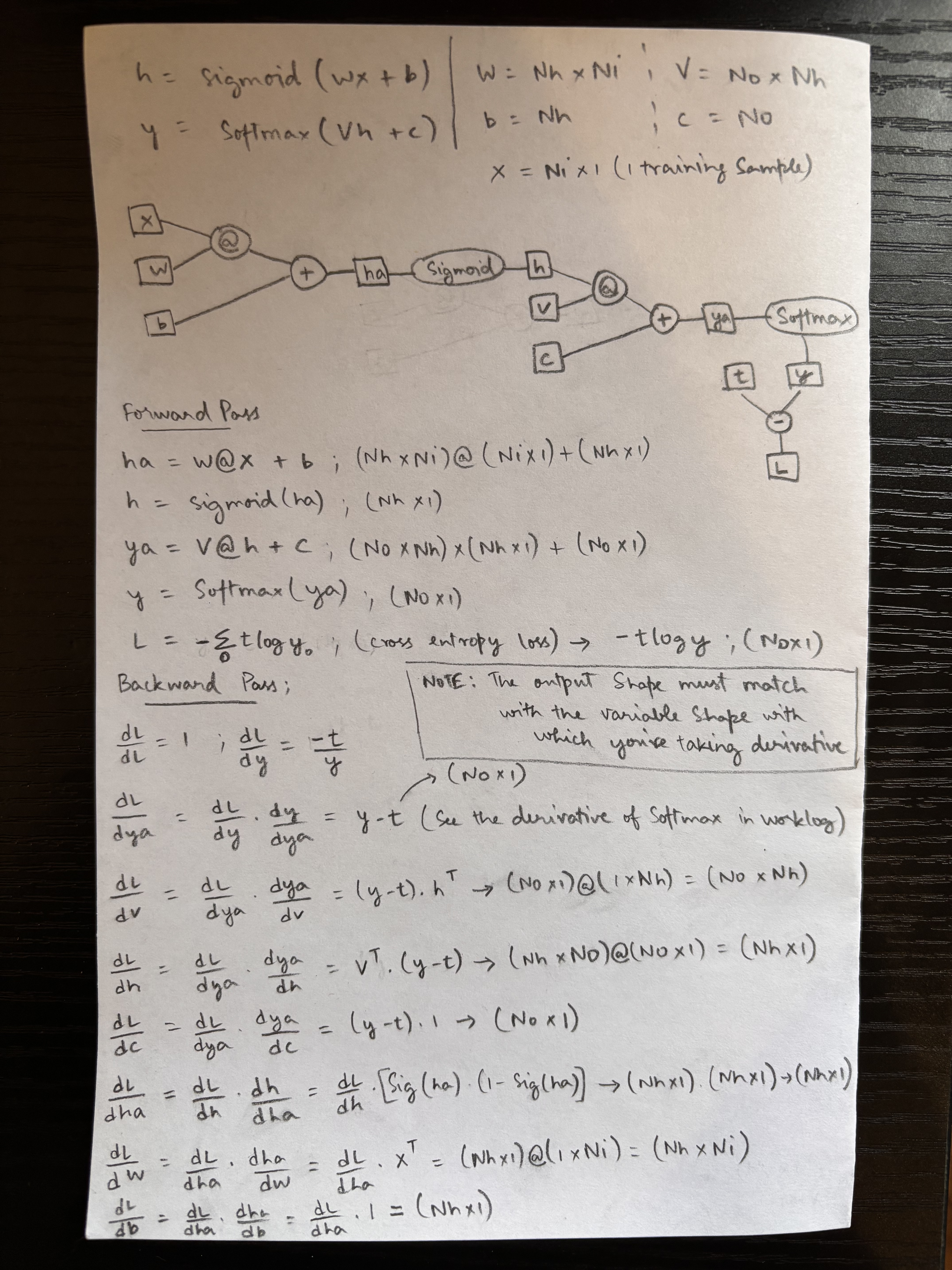
Forward and Backward Pass Using einsum (Single-Sample MLP)
Model
- Hidden layer: \(h = \sigma(Wx + b)\)
- Output layer: \(y = \text{softmax}(Vh + c)\)
Index Convention
- \(i\): input dimension index, \(i = 1 \dots N_i\)
- \(h\): hidden dimension index, \(h = 1 \dots N_h\)
- \(o\): output dimension index, \(o = 1 \dots N_o\)
Forward Pass
Hidden pre-activation
\[h_{a,h} = \sum_i W_{h i} x_i + b_h\]ha = np.einsum("hi,i->h", W, x) + b
Hidden activation (sigmoid)
\[h_h = \sigma(h_{a,h})\]h = sigmoid(ha)
Output pre-activation
\[y_{a,o} = \sum_h V_{o h} h_h + c_o\]ya = np.einsum("oh,h->o", V, h) + c
Output activation (softmax)
\[y_o = \frac{e^{y_{a,o}}}{\sum_{o'} e^{y_{a,o'}}}\]y = softmax(ya)
Loss (Cross-Entropy)
\[L = -\sum_o t_o \log y_o\]Backward Pass
Gradient w.r.t. output pre-activation
\[\frac{\partial L}{\partial y_{a,o}} = y_o - t_o\]dLdya = y - t
Gradient w.r.t. output weights \(V\)
\[\frac{\partial L}{\partial V_{o h}} = (y_o - t_o) h_h\]dLdV = np.einsum("o,h->oh", dLdya, h)
Gradient w.r.t. output bias \(c\)
dLdc = dLdya
Gradient w.r.t. hidden activations
\[\frac{\partial L}{\partial h_h} = \sum_o V_{o h} (y_o - t_o)\]dLdh = np.einsum("oh,o->h", V, dLdya)
Gradient w.r.t. hidden pre-activations
\[\frac{\partial L}{\partial h_{a,h}} = \frac{\partial L}{\partial h_h} \cdot \sigma(h_{a,h})(1 - \sigma(h_{a,h}))\]dLdha = dLdh * sigmoidgrad(ha)
Gradient w.r.t. input weights \(W\)
\[\frac{\partial L}{\partial W_{h i}} = \frac{\partial L}{\partial h_{a,h}} x_i\]dLdW = np.einsum("h,i->hi", dLdha, x)
Gradient w.r.t. hidden bias \(b\)
dLdb = dLdha
Summary;
- All operations are tensor contractions.
-
einsummakes index flow explicit. - Backprop through linear layers reduces to transposes and outer products.
Code:
import numpy as np
# Dimensions
Ni = 784 # Input dimension (e.g., MNIST)
Nh = 500 # Hidden dimension
No = 10 # Output dimension (e.g., 10 classes)
# Initialize parameters
W = np.random.randn(Nh, Ni) * 0.01
b = np.zeros(Nh)
V = np.random.randn(No, Nh) * 0.01
c = np.zeros(No)
# Single training example
x = np.random.randn(Ni) # Input vector
t = np.zeros(No) # One-hot target
t[3] = 1 # Example: class 3
# --- Forward Pass ---
ha = np.einsum("hi,i->h", W, x) + b # (Nh,)
h = 1 / (1 + np.exp(-ha)) # (Nh,)
ya = np.einsum("oh,h->o", V, h) + c # (No,)
# Softmax with numerical stability
ya_max = np.max(ya)
exp_ya = np.exp(ya - ya_max)
y = exp_ya / np.sum(exp_ya) # (No,)
# Loss
L = -np.sum(t * np.log(y + 1e-8))
# --- Backward Pass ---
dL_dya = y - t # (No,)
dL_dV = np.einsum("o,h->oh", dL_dya, h) # (No, Nh)
dL_dc = dL_dya # (No,)
dL_dh = np.einsum("oh,o->h", V, dL_dya) # (Nh,)
sigmoid_grad = h * (1 - h) # (Nh,)
dL_dha = dL_dh * sigmoid_grad # (Nh,)
dL_dW = np.einsum("h,i->hi", dL_dha, x) # (Nh, Ni)
dL_db = dL_dha # (Nh,)
# --- Update parameters (gradient descent) ---
learning_rate = 0.01
W -= learning_rate * dL_dW
b -= learning_rate * dL_db
V -= learning_rate * dL_dV
c -= learning_rate * dL_dc
Question:
When computing
\(\frac{\partial L}{\partial h} = V^\top \frac{\partial L}{\partial y_a},\) using einsum, does einsum implicitly transpose tensors?
Should I change the index order (e.g. use "ho" instead of "oh") to represent the transpose?
Answer:
No. einsum never performs implicit transposes.
All transposes must be expressed explicitly through index labels, not by rearranging axes incorrectly.
In this example, the weight matrix is defined as:
\[V \in \mathbb{R}^{N_o \times N_h}, \quad \text{i.e. } V_{o h}.\]The correct derivative is:
\[\frac{\partial L}{\partial h_h} = \sum_o V_{o h} \frac{\partial L}{\partial y_{a,o}}.\]This maps directly to the following einsum:
dL_dh = np.einsum("oh, o->h", V, dL_dya)